PaleoDeepDive: A system walkthrough
Here, we walk through the codebase that powers PaleoDeepDive, an application built upon the DeepDive machine reading infrastructure. The code can be found here, and one example data dump can be found here.
Prerequisite
To fully understand PaleoDeepDive, it is useful to first go through the online tutorial for DeepDive, which provides an overview of basic concepts and an example of how relations between people, locations, and organizations are inferred. In this document, you will see how similar approaches can be applied to paleontology in order to extract relations between biological taxa, geological rock formations, geographic locations, and geological time intervals.
High-level picture of the application
The goal of PaleoDeepDive is to build a factor graph, like the following, and then estiamte the probabilities associated with each variable and relation. A more detailed description of factor graphs and their use in the PaleoDeepDive can be found in our technical report.
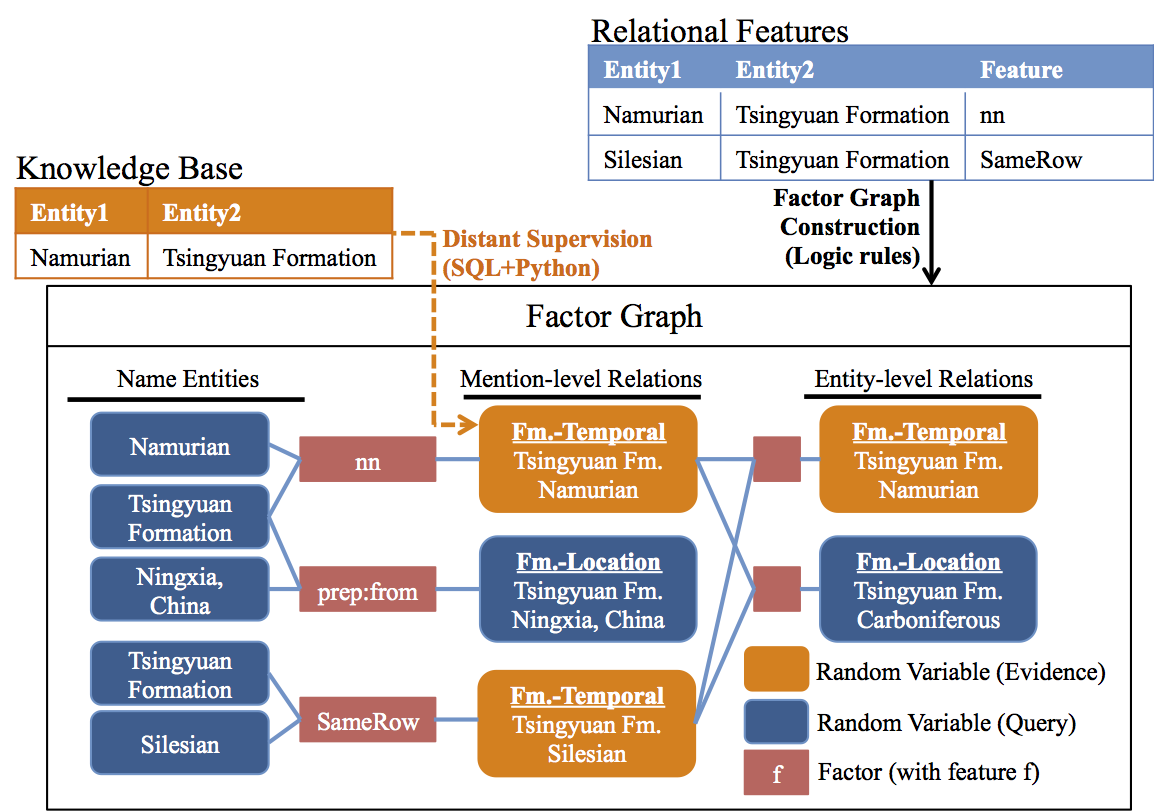
Illustration of the factor graph built in this walkthrough.
To produce this factor graph, we use the following relations that contain random variables.
- Name Entities
entity_formationentity_taxonentity_locationentity_temporal
- Mention-level Relations
relation_formationrelation_formationtemporalrelation_formationlocationrelation_taxonomy
- Entity-level Relations
relation_formation_globalrelation_formationtemporal_globalrelation_formationlocation_globalrelation_taxonomy_global
Contents
Input walkthrough
There are four tables as input, namely (1) sentences (individual sentences within each document) (2) ddtables (tables in PDF documents parsed by existing tools),
(3) layout_align (document layout and formatting information), and (4) layout_fonts (text fonts). These four tables contain
the raw input to PaleoDeepDive, and we will walk through each of them.
Table sentences
The sentences table contains the main text of publications and their
corresponding NLP markups (for more information on NLP).
An example row of this table is as follows.
docid : JOURNAL_106681
sentid : 8
wordidxs : {1,2,3,4,5,6,7,8,9,10,11}
words : {Edzf,U,",",Ciudad,Universitaria,",",Morelia,",",Michoacdn,",",México}
poses : {NNP,NNP,",",NNP,NNP,",",NNP,",",NNP,",",NNP}
ners : {O,O,O,ORGANIZATION,ORGANIZATION,O,LOCATION,O,PERSON,O,PERSON}
lemmas : {Edzf,U,",",Ciudad,Universitaria,",",Morelia,",",Michoacdn,",",México}
dep_paths : {nn,"null",punct,nn,conj,punct,conj,punct,conj,punct,appos}
dep_parents : {2,0,2,5,2,5,5,7,7,2,2}
bounding_boxes : {"[p1l809t1151r871b1181],","[p1l888t1152r912b1178],", ...
This row encodes the sentence
Edzf U, Ciudad Universitaria, Morelia, Michoacdn, México
The columns wordidxs, words, poses, ners, lemmas, dep_paths, and dep_parents are
defined by the NLP software and are consistent with our tutorial. The column
bounding_boxes contains a list of strings, one for each word. For each word,
the string defines a bounding box containing that word in the PDF. As an example, p1l809t1151r871b1181, defines a box on page 1, left margin 809, top margin 1151, right margin 871, and bottom margin 1181 (units are in pixels).
The contents of this table can be used to identify strings and the parts of speech those strings represent, which in turn can be used to define features.
Table ddtables
The ddtables table contains tables in the original PDF document that are parsed
by existing tools (e.g., pdf2table) and our pre-processing scripts. The schema of this table is
as follows.
| id | docid | tableid | type | sentid |
|---|---|---|---|---|
| JOURNAL_102293 | TABLE1DOCJOURNAL102293 | CAPTION | 156 | |
| JOURNAL_102293 | TABLE1DOCJOURNAL102293 | CAPTION | 157 | |
| JOURNAL_102293 | TABLE1DOCJOURNAL102293 | CONTENT | 158 | |
| JOURNAL_102293 | TABLE1DOCJOURNAL102293 | CONTENT | 159 | |
| JOURNAL_102293 | TABLE1DOCJOURNAL102293 | CONTENT | 160 | |
| JOURNAL_102293 | TABLE1DOCJOURNAL102293 | CONTENT | 161 | |
| JOURNAL_102293 | TABLE1DOCJOURNAL102293 | CONTENT | 162 |
These seven rows define a table with ID TABLE_1_DOC_JOURNAL_102293, where the caption consists of sentences with IDs 156 and 157, and content that consists of sentences with IDs 158-162.
Table layout_align
The layout_align table contains layout information in the original document
that is produced by OCR tools and our pre-processing scripts. The schema of this table is as follows.
docid : JOURNAL_100086
property : CENTERED
pageid : 78
left_margin : 1442
top_margin : 1927
right_margin : 2398
bottom_margin : 1960
content : dential canal systems creates recurrent problems in dis-
These eight columns define a bounding box in the PDF document that is ''centered''
in the PDF. The specification of the bounding box is consistent with the
''bounding_boxes'' column of the sentences table.
Table layout_fonts
The layout_fonts table contains the font information of the PDF document produced by OCR tools. The schema
of this table is as follows.
docid : JOURNAL_105969
property : SPECFONT
pageid : 6
left_margin : 1313
top_margin : 698
right_margin : 1856
bottom_margin : 737
content : ("Pseudocetorhinus pickfordi",
Similar to the table layout_align, this row defines a bounding box that
contains a special font.
DeepDive program
Feature extractors
We illustrate the feature extractors in PaleoDeepDive by two examples. First, a simple example that extracts entity-mentions for temporal intervals; Second, a more sophisticated extractors that extract geological rock formations.
The simplest extractor in PaleoDeepDive
We start from the simplest extractor in PaleoDeepDive. The task is
to populate the relation entity_temporal, where each row
is one entity mention for temporal intervals.
id :
docid : JOURNAL_105771
type : INTERVAL
eid : TIME_DOC_JOURNAL_105771_1676_19_19
entity : Permian|298.90000|252.17000
prov : {1676,19,19,Permian}
is_correct :
For this example row, the column eid is a distinct entity mention ID, the column entity is the entity name for temporal interval (in this example, Permian, 298.90000 M.A.-252.17000 M.A.). The column prov contains information to trace back this mention to the original document, in this example, it means that this mention starts from the 19th word and ends at the 19th words of sentence 1676 in the document JOURNAL_105771.
To extract this relation, the schema of the extractor in deepdive.conf is:
ext_entity_temporal_local : {
output_relation : "entity_temporal"
input : """
SELECT docid as docid,
sentid as sentid,
wordidxs as wordidxs,
words as words,
poses as poses,
ners as ners,
lemmas as lemmas,
dep_paths as dep_paths,
dep_parents as dep_parents,
bounding_boxes as bounding_boxes
FROM sentences
"""
udf : ${PALEO_HOME}"/udf/ext_temporal_local.py"
parallelism : 1
}
If you have difficulty in understanding this syntax, please refer to our more general tutorial for DeepDive first. This extractor goes through the sentence table, and for each sentence, it executes the extractor in ext_temporal_local.py.
The python script is as follows.
1 #! /usr/bin/env python
2 from lib import dd as ddlib
3 import re, sys, json
4
5 dict_intervals = {}
6 for l in open(ddlib.BASE_FOLDER + '/dicts/intervals.tsv', 'r'):
7 (begin, end, name) = l.rstrip().split('\t')
8 if name.startswith('Cryptic'): continue
9 dict_intervals[name.lower()] = name + '|' + begin + '|' + end
10 va = name.lower().replace('late ', 'upper ').replace('early ', 'lower')
11 if va != name.lower():
12 dict_intervals[va] = name + '|' + begin + '|' + end
13
14 MAXPHRASELEN = 3
15 for _row in sys.stdin:
16 row = json.loads(_row)
17 docid = row["docid"]
18 sentid = row["sentid"]
19 wordidxs = row["wordidxs"]
20 words = row["words"]
21 poses = row["poses"]
22 ners = row["ners"]
23 lemmas = row["lemmas"]
24 dep_paths = row["dep_paths"]
25 dep_parents = row["dep_parents"]
26 bounding_boxes = row["bounding_boxes"]
27
28 history = {}
29 for start in range(0, len(words)):
30 for end in reversed(range(start + 1, min(len(words), start + 1 + MAXPHRASELEN))):
31
32 if start in history or end in history: continue
33
34 phrase = " ".join(words[start:end])
35
36 if phrase.lower() in dict_intervals:
37
38 eid = "TIME_DOC_" + docid + "_%s_%d_%d" % (sentid, start, end-1)
39 prov = [sentid, "%d"%start, "%d"%(end-1), phrase]
40 name = dict_intervals[phrase.lower()]
41 print json.dumps({"docid":docid, "type":"INTERVAL", "eid":eid, "entity": name, "prov":prov, "is_correct":None})
42
43 for i in range(start, end):
44 history[i]=1
This script contains three components.
- Line 5 - 12: Load a dictionary. One example entry of this dictionary is
{"permian": "Permian|298.90000|252.17000"}. The file ''intervals.tsv'' contains 1K intervals from PaleoDB and Macrostrat. In this extractor, we do not attempt to discover new temporal intervals; however, we do discover new instances for other types of entities, e.g., taxon. - Line 15 - 26: Load the input of this extractor into corresponding Python objects.
Line 28 - 44: Enumerate phrases with up to 3 words, match it with the dictionary, and output an entity mention if it matches. One example output is
{"docid":"JOURNAL_105771", "type":"INTERVAL", "eid":"TIME_DOC_JOURNAL_105771_1676_19_19", "entity":"Permian|298.90000|252.17000", "prov":{"1676","19","19","Permian"}, "is_correct":None}
Note that this example output will produce the example tuple we just show for the table entity_temporal.
A more sophisticated extractor
Here, the goal is to
extract the entity-mention for formations (table entity_formation). One example tuple
in this table is:
id :
docid : JOURNAL_105815
type : shale
eid : ROCK_DOC_JOURNAL_105815_1505_0_1
entity : pierre shale
prov : {1505,0,1,"Pierre Shale"}
is_correct :
This table has a similar structure to the table entity_temporal, above. The extractor attempts to encode the following rule:
If we extracted ''Pierre Shale'' as a formation entity mention candidate, then instances of the word ''Pierre'' in the same document are also a formation entity mention candidate.
To encode this rule, we create an extractor called ext_entity_formation_global
that will be run after an extractor called ext_entity_formation_local
populates the entity_formation with entity mentions like ''Pierre Shale''. The
extractor ext_entity_formation_global is defined as:
1 ext_entity_formation_global : {
2 output_relation : "entity_formation"
3 input : """
4 WITH local_entity_names AS (
5 SELECT docid, array_agg(entity) AS entities, array_agg(type) AS types
6 FROM entity_formation
7 GROUP BY docid
8 )
9 SELECT t0.docid as docid,
10 array_agg((t0.sentid) ORDER BY t0.sentid::int) as sentids,
11 array_stack_int((t0.wordidxs || 0) ORDER BY t0.sentid::int) as wordidxs,
12 array_stack_text((t0.words) ORDER BY t0.sentid::int) as words,
13 array_stack_text((t0.poses) ORDER BY t0.sentid::int) as poses,
14 array_stack_text((t0.ners) ORDER BY t0.sentid::int) as ners,
15 array_stack_text((t0.lemmas) ORDER BY t0.sentid::int) as lemmas,
16 array_stack_text((t0.dep_paths) ORDER BY t0.sentid::int) as dep_paths,
17 array_stack_int((t0.dep_parents) ORDER BY t0.sentid::int) as dep_parents,
18 array_stack_text((t0.bounding_boxes) ORDER BY t0.sentid::int) as bounding_boxes,
19 max(t1.entities) as local_entities,
20 max(t1.types) as local_entity_types
21 FROM sentences t0, local_entity_names t1
22 WHERE t0.docid = t1.docid
23 GROUP BY t0.docid
24 """
25 udf : ${PALEO_HOME}"/udf/ext_formation_global.py"
26 dependencies : ["ext_entity_formation_local"]
27 parallelism : 1
28 }
The SQL query above does two things. First, the WITH clause (line 4-8) defines a relation that contains all existing entity mention candidates. Second, the SELECT clause (line 9-23) defines a relation in which each row contains all sentences in the same document and the corresponding entity mentions.
Distant Supervision
One special class of feature extractors are those used for distant supervision, which is a key technique that we used to produce training labels.
Here, the relation relation_formationtemporal is used as an example.
The feature extractor that we used to distantly supervise the relation
relation_formationtemporal is as follows:
ext_relation_variable_formationtemporal :{
output_relation : "relation_formationtemporal"
input : """
SELECT DISTINCT docid AS docid,
type AS type,
eid1 AS eid1,
eid2 AS eid2,
entity1 AS entity1,
entity2 AS entity2
FROM relation_candidates
WHERE type = 'FORMATIONINTERVAL'
"""
udf : ${PALEO_HOME}"/udf/supervise_formationtemporal.py"
dependencies : ["ext_relation_same_sent", "ext_relation_section_header","ext_relation_from_table"]
parallelism : 1
}
More concretely, the SQL query that this extractor uses returns tuples, such as:
docid : JOURNAL_105815
type : FORMATIONINTERVAL
eid1 : ROCK_DOC_JOURNAL_105815_1289_49_49
eid2 : TIME_DOC_JOURNAL_105815_1289_4_4
entity1 : park formation
entity2 : Precambrian|4000.0000|542.0000
These are candidate relation mentions between entity mentions. The tuple that we want as output is:
docid : JOURNAL_105815
type : FORMATIONINTERVAL
eid1 : ROCK_DOC_JOURNAL_105815_1289_49_49
eid2 : TIME_DOC_JOURNAL_105815_1289_4_4
entity1 : park formation
entity2 : Precambrian|4000.0000|542.0000
is_correct : NULL/True/False
Note that the only difference is the additino of the is_correct column, where
True means a positive example, False means a negative example, and NULL
means that we do not know.
The Python function supervise_formationtemporal.py is used to produce this type of transformation on input tuples.
1 #! /usr/bin/env pypy
2
3 from lib import dd as ddlib
4 import re
5 import sys
6 import json
7
8 kb_formation_temporal = {}
9 for l in open(ddlib.BASE_FOLDER + "/dicts/macrostrat_supervision.tsv"):
10 (name1, n1, n2, n3, n4) = l.split('\t')
11 name1 = name1.replace(' Fm', '').replace(' Mbr', '').replace(' Gp', '')
12 n1, n2, n3, n4 = float(n1), float(n2), float(n3), float(n4.rstrip())
13
14 for rock in [name1.lower(), name1.lower() + " formation", name1.lower() + " member"]:
15 if rock not in kb_formation_temporal:
16 kb_formation_temporal[rock] = {}
17 kb_formation_temporal[rock][(min(n1, n2, n3, n4), max(n1, n2, n3, n4))] = 1
18
19 for _row in sys.stdin:
20 row = json.loads(_row)
21 docid = row["docid"]
22 type = row["type"]
23 eid1 = row["eid1"]
24 eid2 = row["eid2"]
25 entity1 = row["entity1"]
26 entity2 = row["entity2"]
27
28 if entity1 in kb_formation_temporal:
29 (name, large, small) = entity2.split('|')
30 large = float(large)
31 small = float(small)
32
33 overlapped = False
34 for (a,b) in kb_formation_temporal[entity1]:
35 if max(b,large) - min(a,small) >= b-a + large-small :
36 donothing = True
37 else:
38 overlapped = True
39
40 print json.dumps({"docid":docid, "type":type, "eid1":eid1, "eid2":eid2, "entity1":entity1, "entity2":entity2, "is_correct":None})
41 if overlapped == True:
42 print json.dumps({"docid":docid, "type":type, "eid1":eid1, "eid2":eid2, "entity1":entity1, "entity2":entity2, "is_correct":True})
43 else:
44 print json.dumps({"docid":docid, "type":type, "eid1":eid1, "eid2":eid2, "entity1":entity1, "entity2":entity2, "is_correct":False})
45 else:
46 print json.dumps({"docid":docid, "type":type, "eid1":eid1, "eid2":eid2, "entity1":entity1, "entity2":entity2, "is_correct":None})
This extractor contains four components.
- Line 8 - 17. Load the dictionary for distant supervision from macrostrat. One entry
for the dictionary kb_formation_temporal is like
{"pierre shale":{(66.0,89.8):1}}. - Line 19 - 26. Load the input tuple from SQL queries.
- Line 28 - 38. Check whether there exists one entry in kb_formation_temporal with the same formation and an overlap temporal interval.
- Line 40 - 46. Produce output tuples based on the result of checking.
- Line 40: Output one tuple with label None no matter what. (Even if we know this tuple is true, we still need to reclassify it based on the learning result to be able to use it to produce inference result. PaleoDeepDive can never directly use the result of distant supervision as inference result.)
- Line 42: If overlap, then output a positive example.
- Line 44: If not overlap, then output a negative example.
- Line 46: If the formation is not in the dictionary, then output None.
Inference Rules
Inferences rules are used to specify the correlation among random variables. Most of these inference rules have a similar form as the one in our tutorial, and here we show two examples.
inference_rule_formation : {
input_query: """
SELECT t0.features,
t1.id as "relation_formation.id",
t1.is_correct as "relation_formation.is_correct"
FROM relation_candidates t0, relation_formation t1
WHERE t0.docid=t1.docid AND t0.eid1=t1.eid1 AND t0.eid2=t1.eid2
"""
function: "Imply(relation_formation.is_correct)"
weight: "?(features)"
}
In this inference rule, predictions are made for random variables in relation_formation,
which derive from relation_candidates. The result
of this SQL query:
features : [SAMESENT PROV=INV:]
relation_formation.id : 7564
relation_formation.is_correct :
The column ''features'' defines the justification for the relation and DeepDive will train a weight for each feature.
A more sophisticated example is as follows:
inference_rule_formationtemporal_global1 : {
input_query: """
SELECT t0.id as "global1.id",
t1.id as "global2.id",
t0.is_correct as "global1.is_correct",
t1.is_correct as "global2.is_correct"
FROM relation_formationtemporal_global t0,
relation_formationtemporal_global t1,
interval_containments t2
WHERE t0.docid=t1.docid AND t0.entity1=t1.entity1 AND t0.entity1=t2.formation
AND t0.entity2=t2.child AND t1.entity2=t2.parent
"""
function: "Imply(global1.is_correct, !global2.is_correct)"
weight: "10"
}
In this inference rule, connections between two random variables ar made, both of which come from
relation_formationtemporal_global. The SQL query returns a pair of
random variables that have the same formation, but different temporal intervals, such that
one contains the other (e.g., Cretaceous and Late Cretaceous). The inference
rule says that if the smaller interval is correct, then we do not want to output
the larger interval (because it is redundant). This rule is a hard rule, meaning that a large weight is assigned a priori.
Output walkthrough
The output of DeepDive takes the form of relations, for example the realtion called relation_formationtemporal_is_correct_inference,
which contains tuple:
id : 4426
docid : JOURNAL_105815
type : FORMATIONINTERVAL
eid1 : ROCK_DOC_JOURNAL_105815_658_27_27
eid2 : TIME_DOC_JOURNAL_105815_658_30_31
entity1 : morrison formation
entity2 : Late Jurassic|161.2000|145.5000
is_correct :
category : 1
expectation : 1
id : 4407
docid : JOURNAL_105815
type : FORMATIONINTERVAL
eid1 : ROCK_DOC_JOURNAL_105815_3356_10_12
eid2 : TIME_DOC_JOURNAL_105815_3356_14_14
entity1 : white river formation
entity2 : Oligocene|33.90000|23.03000
is_correct :
category : 1
expectation : 1
Compared with relation_formationtemporal, there are two new columns
in the relation relation_formationtemporal_is_correct_inference, namely
(1) category and (2) expectation. This means that after inference, DeepDive
calculates the expectation of the following function: if the value of the random variable equals the category, then 1, otherwise 0. In this case, one can conceive of the expectation as the probability of the corresponding random variable being true.

